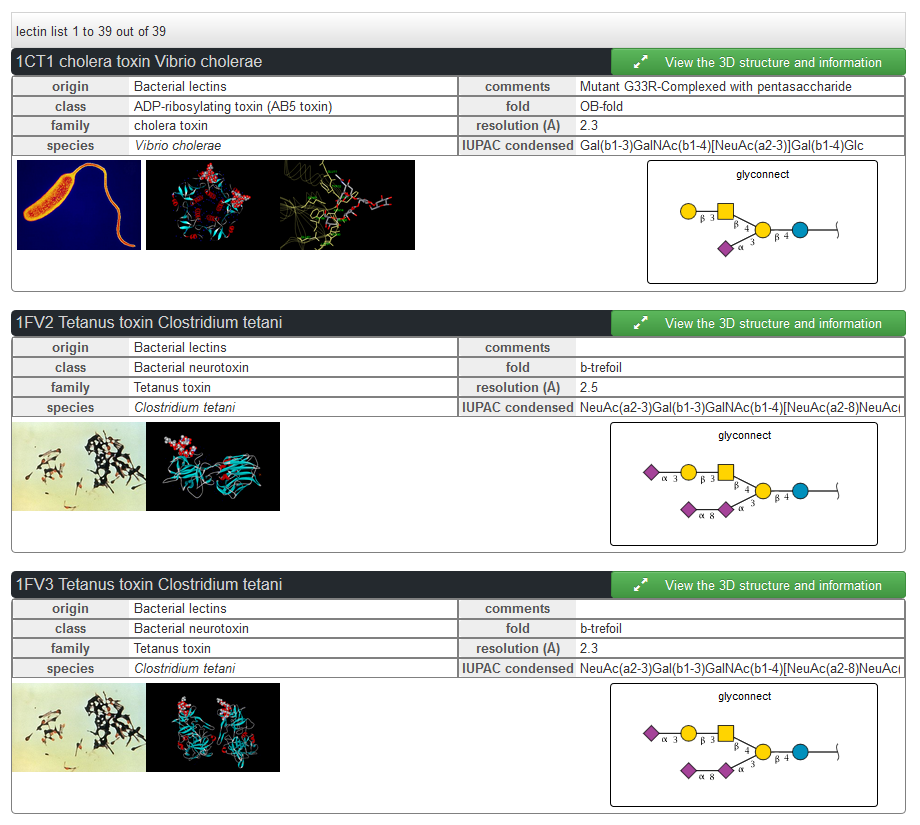
The UniLectin3D database contains the three-dimensional structures of Lectins that have been crystallized with or without their carbohydrate (glycans) ligands and non-carbohydrate ligands (see Background for detailed information). The following options for searching are available: 1. Keywords 2. Kingdom order, carbohydrate binding site class and species family 3. Monosaccharide and associate IUPAC sequence 4. Search by binding site fold 5. Search by multiple criteria Once selected, lectins can be explored (and their features pictured and downloaded): 6. Sequence (with the UniProt AC) 7. Structure (with the PDB ID) For each lectins a detailed page is available with 3D visualization, interactions and links to external databases.

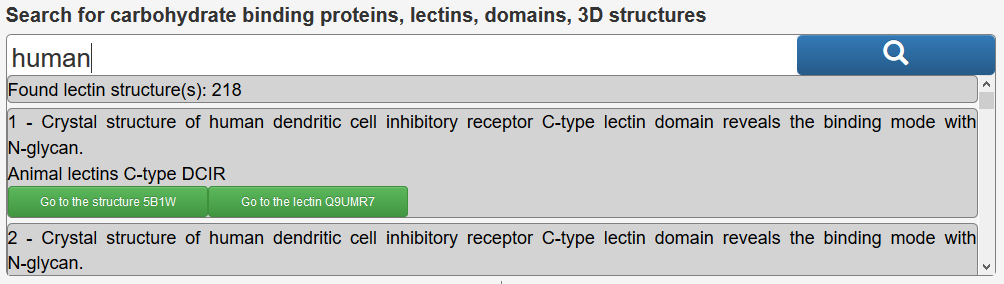
The search can be performed by entering keywords by textual input : i.e. human, PDB code, UniProt accession number, lectin name, type of domains, fragment of glycan sequence, or textual fragments of the title of a publication.,….i.e : human, propeller, 1TL2 (PDB), Q47200 (UniProt), GalNAc, Lewis,…. Example: Entering “human” in the search box, will return the following.
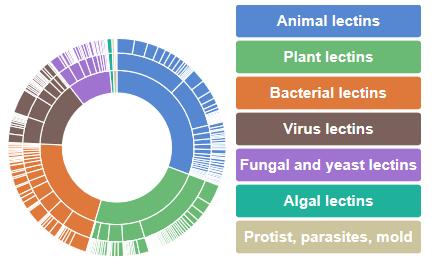
The inner circle corresponds to the kingdom orders along with their respective percentage of occurrence. For a given kingdom order, (for example animal lectins) the carbohydrate binding site class appears in the central section (for example galectins) whereas the species families can be browsed on the outer section (for example galectin 3).
The taxonomic tree also allows to explore the classification with the tree leaves which can be clicked to expand the tree and access subcategories
Glycan are described by their monosaccharide content and encoded in IUPAC condensed format (nomenclature of carbohydrates).
Ex. Gal(b1-4)GlcNAc(b1-2)Man(a1-3)[GlcNAc(b1-2)Man(a1-6)]Man
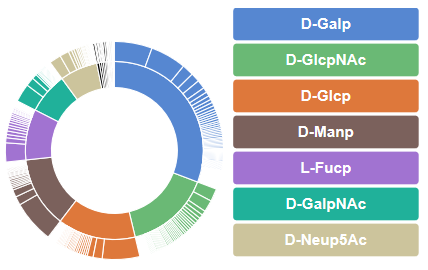
The inner circle correspond to the monosaccharide that compose the carbohydrates chains interacting with the lectins, together with their respective percentage of occurrence. The outer circle correspond to the carbohydrate chains. For a given monosaccharide (D-Galp for example) appears the number of occurrences of glycans containing the selected monosaccharide (for example 29 in the case of Gal(b1-3)GalNAc).
Clicking on a selected glycan (for example Gal(b1-3)GalNAc) lists the 3D structures of lectins complexed with this carbohydrate.
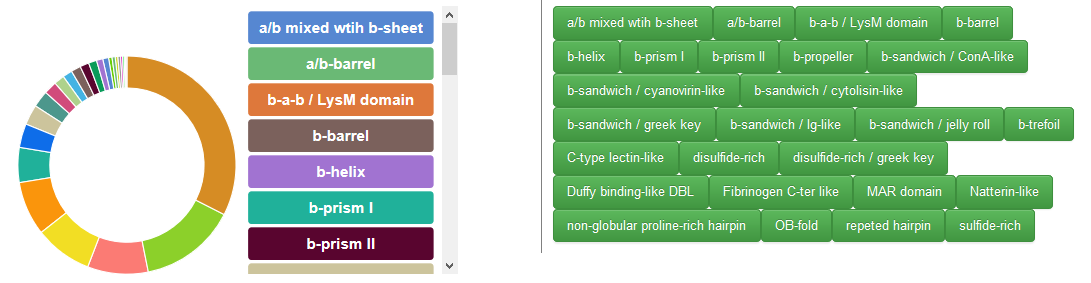
The circle correspond to the fold identified in the crystal structures of lectins used to interact with the carbohydrate chain, together with their respective percentage of occurrence. Buttons are also displayed to access the lectin with the respective fold.

This advanced search option offers a range of criteria to be selected in a combined fashion to search throughout the whole content of the database. The research interface provides multiples criteria to select specific lectins or structures. For every structure multiple manually curated information are available, mostly about the associate carbohydrate.
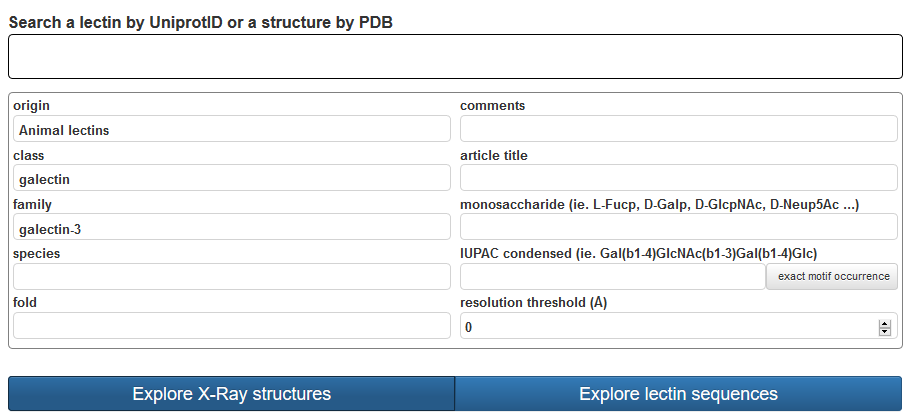
The advanced search offers selection of criteria. Lectin can be searched by structure or by sequence family with the support of drop-down lists. The classification of lectins (Origin, Class, Family) also provides several search criteria. Other criteria pertain to the nature of the fold and taxonomic details of the lectin. Keywords from the title in a reference article can also be searched. A unique feature is the search of fragments of glycan ligands that will output lectins interacting with a given carbohydrate. Finally, a cutoff on the resolution (Å) of the X-ray structure can be used as a filter for selecting of high-quality data. UniLectin3D allows precise taxonomic search for all lectins that have been structurally characterized in a given organism. Another key feature is the option to search lectin structures with oligosaccharide motifs that are complexed within the binding sites.
The resolution criteria relates to the resolution of the structure determination (High numeric values of resolution, such as 4 Å, mean poor resolution, while low numeric values, such as 1.5 Å, mean good resolution. 2.05 Å is the median resolution for X-ray crystallographic results in the Protein Data Bank). Whenever the resolution is set to 0, the structures are not filtered.
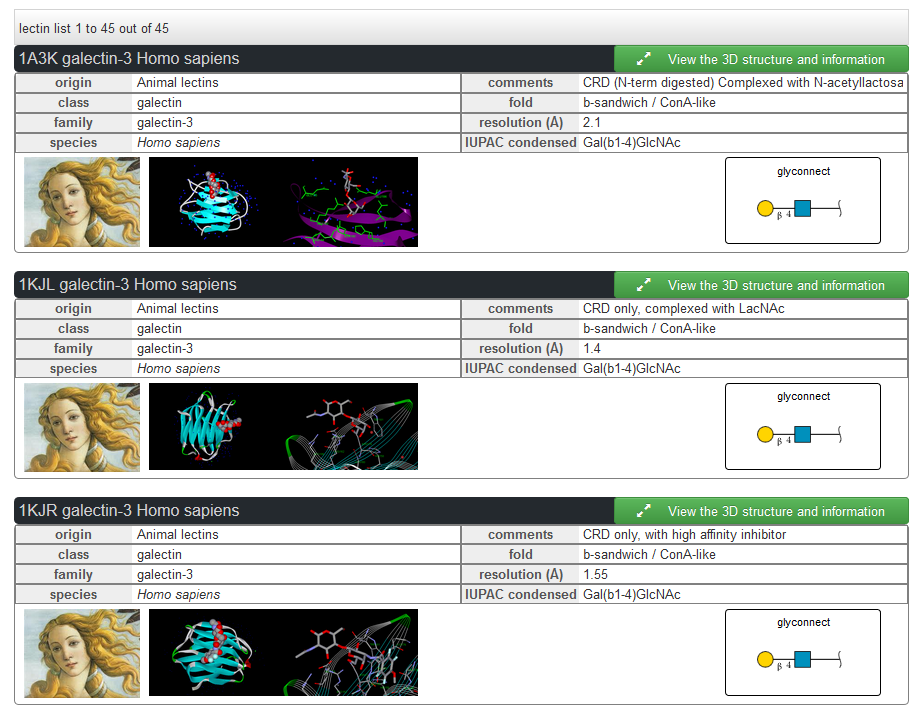
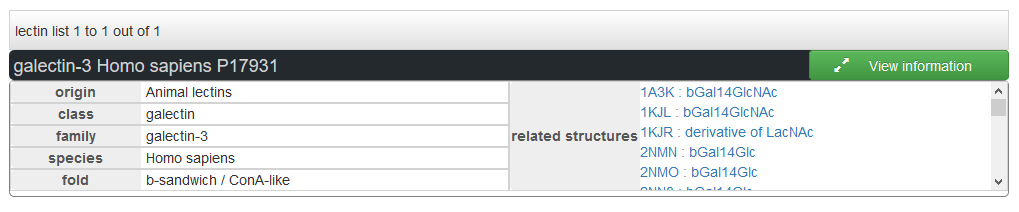
The lectins can be explored by protein uniprot AC. One lectin can be related with multiples PDB structures, which are displayed in the results together with the ligand.
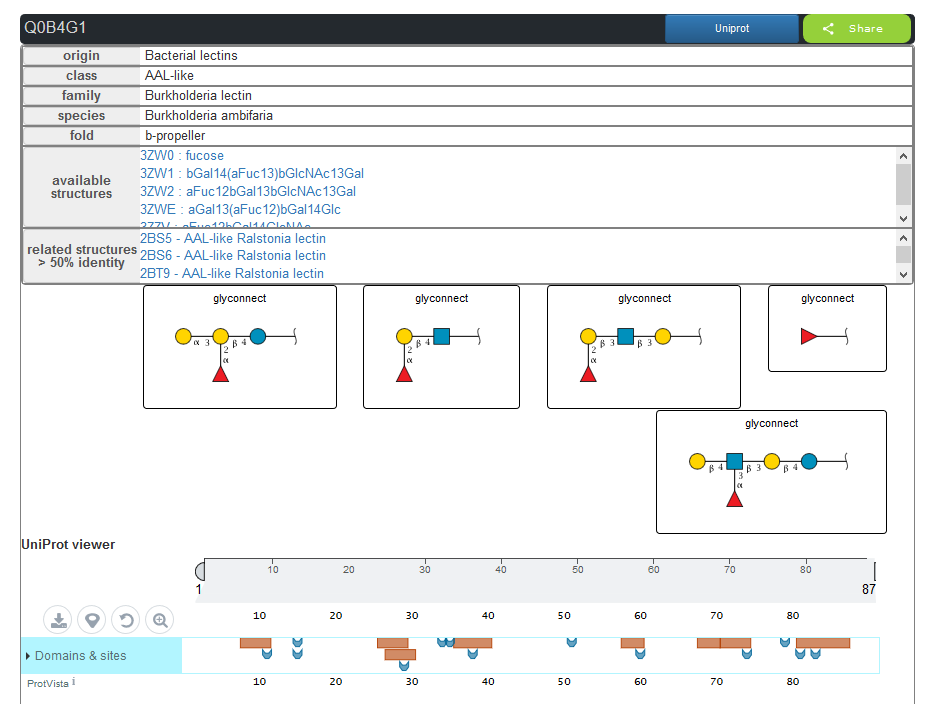
The lectins can be explored by struture. Each structure is related to a protein with an ID uniprot. For each structure the other structure from the same protein are proposed in the results.
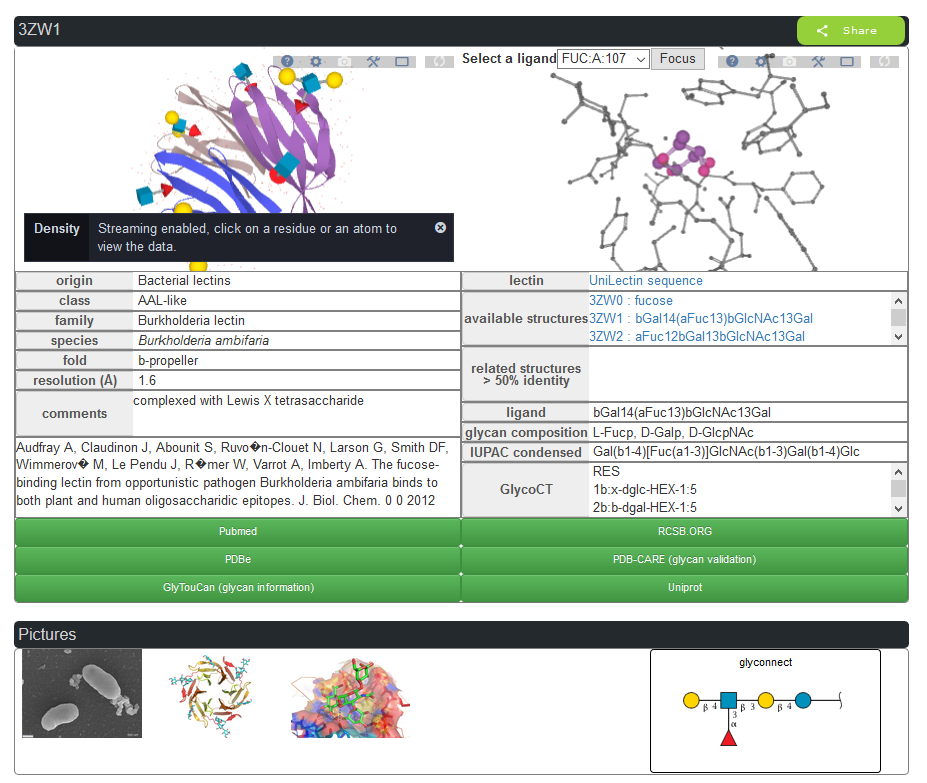
Information can be obtained for the protein partner, the glycan partner, and details of their interactions using the Protein-Ligand Interaction Profiler (PLIP) server. Special care was devoted to the description of the bound glycan ligands with the use of simple graphical representation and numerical format for cross-linking to other databases in glycoscience. We conceive the architecture and navigation tools to extend the search to all organisms, as well as to search for all oligosaccharide epitopes complexed within specified binding sites.
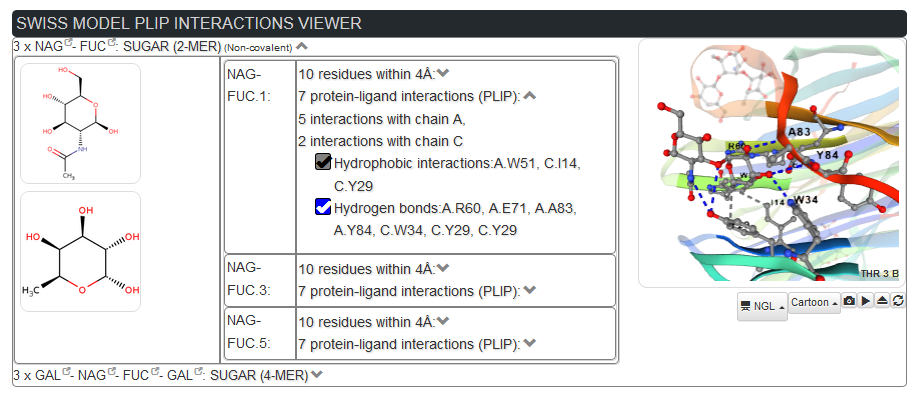
PLIP allows to visualize in details the interactions on the 3D structure, specific interactions types, and the surounding residues
The PDBe functionaol domain viewer provides information on the glycosilated sites, functional domain, carbohydrates binding sites
The UniLectin3D project gathers structural information on lectins, which are glycan-binding proteins from all origins, along with their interactions with carbohydrate ligands. Among the proteins that interact non-covalently with carbohydrates, lectins bind mono- and oligosaccharides reversibly and specifically while displaying no catalytic or immunological activity. Lectins are oligomeric proteins that can specifically recognize carbohydrates, which as per present knowledge act as macromolecular tools to decipher sugar-encoded messages. Those complex carbohydrates (also referred to as glycans), both in the form of single molecules, or bound to proteins and lipids, are the most abundant class of biomolecules. They are increasingly being implicated in human health and environmental issues. Complex carbohydrates are built for high-density bio-coding, which is at par with proteins and nucleic acids, The information carried by glycans is encoded by their 3D-structure and sometimes by their dynamics. Consequently, the structural characterization of lectin interactions, as a reader of the glyco-code is essential.
Lectins are still poorly classified and annotated, and since their functions are based on recognition, we use their 3D-structures as the foundation of project. UniLectin3D is a curated database with a classification proposed on origin and fold with association of literature and functional data such as known specificity. The content of UniLectin3D is centered on 3-dimensional data, using PDB information, with an appropriate curation of the glycan topology. It provides a family-based classification and cross-links to specialized glyco-related databases. In particular, each carbohydrate ligand can be seen (upon one click) as part of the full carbohydrate structures. Finally, the 3D visualization of contacts between the lectin and the ligand, is visualized via the Protein-Ligand Interaction Profiler (PLIP) server. The introduction of such a feature is likely to meet the expectations of lectin specialists. The UniLectin3D contains 1740 lectin structures that have been manually curated; this corresponds to 428 different lectins (as of 2018-08-29). Bibliographic entries cover the 766 published articles describing at least one structure. The first classification level, referred to as origins, separates the lectins into seven different classes, which correspond to the main domains of the living kingdom. The second level orders the lectins according to the protein fold into 75 classes. The third level separates the lectins according to their species in 309 families.
Among the 1740 3D structures, 1120 occur as complexed with glycans. The most commonly observed monosaccharides are as follows: Galactose (Gal) 34%(600); N-Acetyl glucosamine (GlcNAc) 17%(302), Glucose (Glc) 15%(275), Mannose (Man) 14%(251), Fucose (Fuc) 10%(179), N-Acetyl galactosamine (GalNAc) 8% (145), sialic acid (Neu5Ac) (134) 7%., but rarer sugars are also observed in complexes with lectin (Rhamnose, Arabinose …). The ligands occur as monosaccharides, but also as oligosaccharides or glycoconjugates. The set of distinct glycan ligands amounts to 188.
 How to cite
How to cite